Complete mitogenomes from near-extinct tribes in North Altai: New insights into Early Siberians linked to Native Americans
Elena B. Starikovskaya1, Azhar M. Nazhmidenova1, Stanislav V. Dryomov1, Sofia A. Shalaurova1, Ilya O. Mazunin1, Anatoly P. Derevianko2, and Rem I. Sukernik*,1
1Laboratory of Human Molecular Genetics, Institute of Molecular and Cellular Biology, SB RAS, Novosibirsk, Russian Federation
2Institute of Archaeology and Ethnography, SB RAS, Novosibirsk, Russian Federation
*Correspondence: sukernik@mcb.nsc.ru
Abstract
North Altai (present-day Altai Republic/Altai Region) is an area evidently central to the large-scale colonization of Siberia. Previous studies, employing ethnographic and mtDNA data as a baseline to interpret archeological manifestations, have displayed the key role of the primordial hunting culture for game originating within the Altai-Sayan taiga region during the pre-Bronze Age. Through a comprehensive dataset of complete mitochondrial genomes (n=142), we identified a clear genetic sub-structure across the Tubalar, Chelkan, Kumandin, and Teleut people, which remain poorly sampled, and these new data greatly enhance our understanding of the peopling of Early Siberia and North America. Although 27.5% of the mitogenomes fall into western Eurasian lineages (e.g., N9a, H8b1, U2e1, U4b1, and U5a1d2b), other lineages found within our samples likely resulted from admixture with Northeastern Paleoasiatic groups with whom Altai-Sayan people assimilated over time. Remarkably, we identified a unique complex B4b1a/B4b1a3a, disclosing a close link between this component of the Tubalar/Chelkan ancestry and Native American B2.
Keywords: Mitochondrial genomes; Early Siberians; Native Americans
Introduction
The Altai-Sayan Mountain system has long been recognized as a strategic area of very high relevance to the geographic center of Eurasia where the earliest Siberians (as already represented by archaeogenetic data from remains like Afontova Gora near the Yenisei river) left their mark in Altai and adjoining regions, and where major prehistoric events occurred during the Neolithic and subsequent periods (Derevianko and Markin 1998). Groups that may have evolved sequestered from one another until the late Neolithic intermingled with seminomadic people who had migrated from the eastern Eurasian steppe, leading to the eventual assimilation of resident hunter-gatherers. Toward the end of the Bronze Age, west-east movements carried pastoralism into intermountain basins in south Siberia (Czaplicka 1918; J. P. Mallory 1997; Anthony and Ringe 2015; Lamnidis et al. 2018).
In our earlier studies, the core of the mtDNA makeup of native people from northeastern Altai was found to consist of mtDNA lineages that apparently were ancient European or at least western Eurasian, but a portion of the Tubalar sample was from founding lineages that might correspond to a sustainable place of refuge in the Altai-Sayan area for groups also giving rise to Native American ancestors (Starikovskaya et al. 2005; Volodko et al. 2008; Sukernik et al. 2012).
In the present report, newly obtained entire mtDNA sequences were integrated with those previously published, with a focus on our new survey of unrelated mitochondrial DNA genomes retained in the Tubalar, Chelkan, Kumandin, and Teleut. Accordingly, we have completed filling gaps in the mtDNA genome diversity which remained vaguely sampled in Indigenous Altaians previously (Dulik et al. 2012). These are the groups that likely formed through a long process of interbreeding between Uralic and Turkic-speaking tribes, as well as the Yenisei Ostyaks (Ket) living on the right bank of Yenisei and its tributaries (Radloff 1882; Aristov 1896).
Results
The North Altaian mtDNA lineage structure is relatively complex, with most lineage segments ultimately claiming descent from lineages of related subgroups. Of 142 mitochondrial genomes listed in Table S1, three A-152! m.16362 (A4 in Sukernik et al. 2012) mtDNA genomes - two Tubalar and one Kumandin - are new samples. Aside from the unique Tubalar B4b1a (AY519494.2) complete mtDNA genome disclosed previously (Starikovskaya et al. 2005; Sukernik et al. 2012), the B4b1a3a mitogenomes assigned to the Tubalar, Chelkan, or Kumandin are newly sequenced. Frequency determination of different mt haplotypes per four populations is presented in Table S2.
Among rare mitogenomes retained in extant Northern Altaians are those that belong to western Eurasian lineages such as I4a identified in a single Chelkan individual. In contrast to that found in Europeans, originally identified as H8 (Achilli et al. 2004; Loogväli et al. 2004), H8b1 embraces three different sub-branches: m.960.4C in Estonia, m.961T>C marking a node assignment of a subset of the sequences (Teleut, Tubalar, Kumandin, or Shor), whereas m.203G>C, m.15221G>A, and m.16344C>T mark the Evenki samples. The updated age of the H8b1 cluster is 4.2 ka, consistent with its pattern of current geographic distribution (Additional file, Fig S1).
Generally, the Tubalar mtDNA samples are haplogroup U mtDNAs assigned to subhaplogroups U2e1, U4a1d, U4b1, and U5a1d2b. A phylogenetic tree of the U4a/U4a1 lineages encompasses ancient mtDNA sequences from those early or "Old Siberians" who underwent pronounced differentiation in the Altai-Sayan region, followed by far-reaching dispersals and subsequent isolation between ancestral and subsequent groups. Accordingly, the U4b1a4 and/or U4b1b1 subtypes found in either Tubalar or Kumandin revise our understanding of the U4b1 sub-haplogroup relationships (Additional file, Fig S2). The uneven distribution of the two sub-haplogroups, with U4b1a4 in Tubalar/Kumandin and U4a1 predominantly found in Ket, evidently comes from the ancestral status of U4a and U4b in the region.
Of the 46 Teleut samples, 11 (23.9%) were found to fall into distinctive mtDNA lineages of either present-day Europeans or west Eurasian origin (e.g., H8b1, with a single instance of N1a1a1a1a). More common haplogroups J and T (sub-branches J1, T1, T2) evidence traces of diffusion events in European prehistory that may have been involved in Late Glacial expansions starting from the Near East before Neolithic times (Fu et al. 2012; Fernandes et al. 2012; Olivieri et al. 2013). The J1 and T1 samples, attested in single localities (Shanda, Bekovo, Noviy Bachat) attributable to the Salair Range region, appear to be unique for modern Teleut. Despite their geographical proximity and tentatively hypothesized shared homeland, quite a few instances of shared East Eurasian mtDNA lineages are confined to Teleut and Kumandin: the high frequency of C5c revealed in both groups. The occurrence of X2e2a1 sequences in a sole Tubalar and three Kumandin samples implies that traces of the European X2e2 sub-branch separated from the Near Eastern X2e root by three mutational steps (3948-12084-13327) represent a portion of relatively recent gene flow toward Altai-Sayan (Sukernik et al. 2012). While the Kumandin mitochondrial genome diversity is distinctive for D4j10 and D4m2a, the Chelkan are distinguished from the rest in that they have haplogroup N9a9, F2b1, and G2a2 mtDNA genomes. Unlike N9b subdivisions restricted to the Primorye region from the Russian Far East (Dryomov et al. 2020), haplogroup N9a9 prevails in the Altai-Sayan region. Remarkably, an apparent N9a variant was recently identified in archaeological remains from the Carpathian Basin (Rusu et al. 2018). The unique D5a2a1-16172! mitogenome samples identifiable in the upper Biya River from the villages of Turochak (MG660520) and Pyzha (EU482378), as well as a sole Todji (MG660771) from Tuva, are also noteworthy.
Pairing modern and ancient mitogenomes refines the ancestral D4b1a2a cluster (Additional file, Figure S3). One of the subclusters, distinguished by m.13759G>A, gives rise to a rare haplotype (EU482305.1) retained in one of the 31 samples, presumably of Yukaghir origin (Volodko et al. 2008; Sukernik et al. 2010). The other node (m.13720C>T) gives rise to two subbranches, D4b1a2a1 and its sister D4b1a2a2. The principal offshoots of the D4b1a2a2 lineage (EU482385 and FJ951487) extend to Mongolia (Derenko et al. 2010). The distinctive sub-lineage D4b1a2a1a1 defined by m.11383T>C and m.14122A>C represents a relatively recent split of D4b1a2a1a extending the range of diversity events during the Holocene and later (Dryomov et al. 2015, 2021). Before the major split and subsequent spread of Inupiaq speakers into northern Alaska and the Canadian Arctic, the age of the entire D4b1a2 cluster is ~10.65 kya.
Discussion
The expansion from Siberia into the Americas (16 to 13.5 kya) is consistent with terrestrial/coastal migrations, and this evidence may explain the absence of existing consensus regarding the routes and timing of the peopling of the Americas (Braje et al. 2018). Derived from contemporary populations and ancient individuals, genomic data confirm that the first Americans originated from Asia and after several population splits moved south of the continental ice sheets that covered Canada sometime between ~17.5 and ~14.6 kya (Potter et al. 2017; Moreno-Mayar et al. 2018). Recent research provides genetic support for a South American settlement before 18 kya (Sepúlveda et al. 2022). A complex palimpsest of dispersal, lineage extinction, and persistent matrilineal connections has led Neel et al. (1994) to the suggestion that the ancestry of the Amerindian is multipartite, derived in part from northeastern Siberian groups (the source of haplogroups A, C, and D) and in part from groups to the south, the source of haplogroup B. Thus, it can be seen (Bisso-Machado and Fagundes 2021) that mtDNA haplogroup B is practically absent in northern North America and the extreme south of South America, as well as in the Amazon. Today, mitochondrial DNA of haplogroup B2, nested within parental 'Asian' B4, is one of the few haplogroups found exclusively among Indigenous peoples of the Americas (Wood et al. 2019). Beringia was a refugium during the height of the last glacial maximum before climate change and the retreat of the ice sheets allowed access to the remainder of the Americas (Potter et al. 2014; Llamas et al. 2016).
Materials and Methods
A set of 296 individuals representing 142 global populations from the Simons Genome Diversity Project (SGDP) (Mallick et al. 2016) ensures Global Analysis exploring Mobile Element Diversity (MEI) from deeply sequenced whole-genome data (Watkins et al. 2020), and in this way defines the geographic locations of all 'Old World' individuals with >1% New World shared ancestry, to calculate a geographic centroid within Asia for the Native American shared ancestry component. The centroid (cross-hatched circle) occurs in Khakassia (53.92 latitude, 90.66 longitude). Using only individuals with substantial New World ancestry (>6.25%) moves this centroid to the northwest into the upper Baikal and Lena River regions (60.47 latitude, 119.08 longitude). The results, based on genome-wide strictly identical-by-descent MEI insertions, provide a robust inference of a southern to southeastern Siberian origin for the primary wave of early migrants that gave rise to most Native Americans today. This signal of shared ancestry probably corresponds to the "First Americans" migration wave, which is one of at least three streams of gene flow from Asia to America (Reich et al. 2012). It is not surprising that the respective centroid occurs in Khakassia, the area bordering the Altai-Sayan region where the Tubalar/Chelkan complex B4b1a/B4b1a3a is particularly located.
Populations and samples
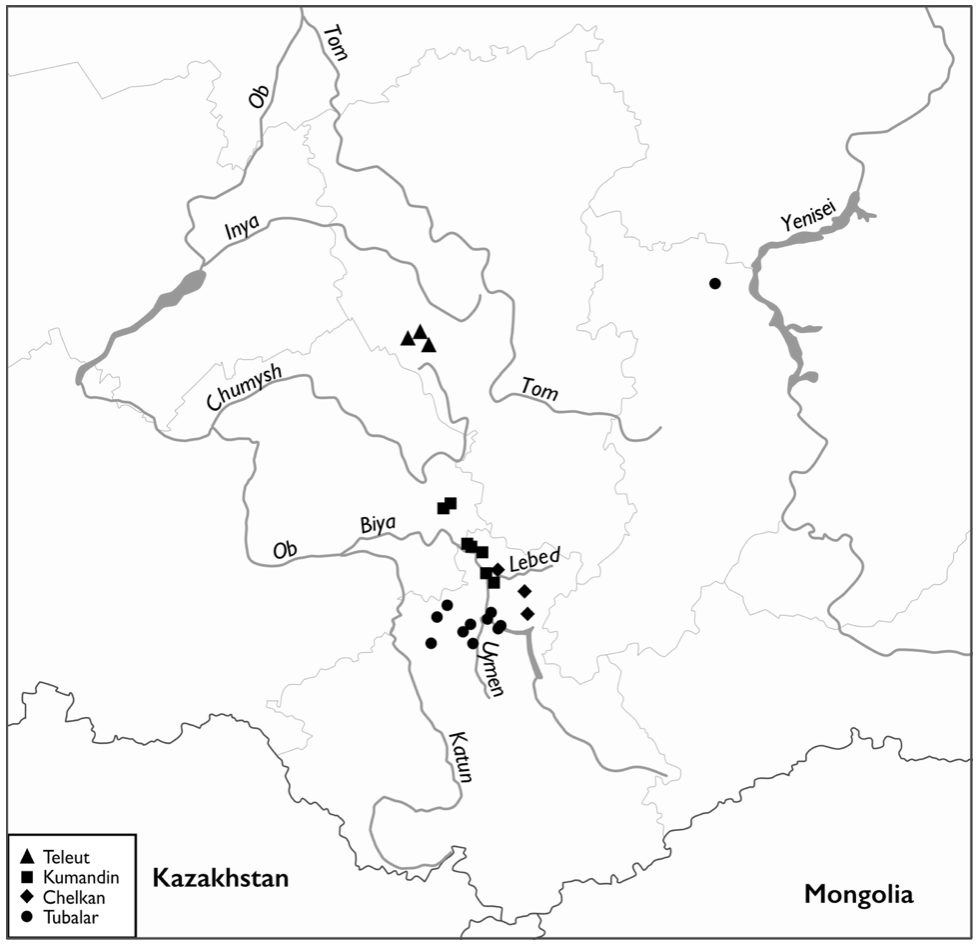
Blood samples were collected from subjects who lacked non-native maternal ancestors verified by senior members of the community for accuracy of the compiled genealogies prior to blood being drawn. Natives with known Russian admixture have been excluded from the study. The sample area is shown in Figure 1, and a brief description of each tribe follows:
Tubalar. The census of 2002 recorded ~1,500 members who are thought to be descendants of some of the earliest inhabitants of the taiga refuge of Altai-Sayan, encompassing the northern coast of Lake Teletskoye, the upper left coast of the Biya River, and the Isha River (the Upper Ob River basin). On the northeastern border of their range, the Tubalar were closely related to the Chelkan tribe that dwindled during the last decades (Levin and Potapov 1964; Potapov 1969).
Chelkan. This subgroup of a few hundred individuals in the middle of the 20th century represents the now-amalgamating remnants of two exogamous lineages dispersed in their traditional territory, encompassing the sources of the Lebed River and Baygol River (Potapov 1969). We selected maternally unrelated samples from 26 elders, born in or derived from tiny settlements: Suronash, Kurmach-Baygol, Chebichen, and Itkuch, the latter two of which no longer exist. Importantly, along with nearby Shor and "Abakan Tatar"/"Kamasintsi" of the former Yeniseysk province, these people are classified as the 'Old Siberians' (Lopatin 1940; Dryomov et al. 2021).
Kumandin. The Kumandin people in the early 17th century lived along the river Charysh, near its confluence with the Ob River, and survived by hunting and working off the taiga as herders and nomads (Lopatin 1940; Potapov 1969). A subsequent relocation to northeastern Altai was driven by their unwillingness to pay tribute to the Russian sovereign (Aristov 1896). The 59 blood samples were collected in 2016-2017, during several field works in the villages of Ozero Kureyevo, Dmitrievka, Shunarak, Turochak, Sankin-ail, Kebezen, and Biyka (Turochak district, Altai Republic), and the villages of Solton and Shatobal (Solton district, Altai Region).
Teleut. Russian documents of the 17th‒18th centuries note the Salair Ridge extending along the northern border of Altai Region as a major center of gathering and population dispersal of Teleut, also described as "White Kalmyk" (Potapov 1969; Funk 2005). A population count of fewer than 1,000 "White Kalmyk" in all was noted in 1897 (All-Russian census). In the Middle Ages, these people comprised thousands of individuals spread over a vast forest-steppe region from the Irtysh River to the Altaian foothills. Some of their subdivisions represented a pastoral people who traditionally subsisted by herding horses, whereas others were pedestrian hunters. "White Kalmyk," "Telengit," and "Telesy" were occasionally used as alternative names for the Teleut, speaking a language of the Altai subgroup of the Kirgiz-Kypchak group in the eastern branch of the Turkic language family (Funk 2005). They are inferred to be related to the Tiele people, a confederation of several seminomadic tribes of different Turkic ethnic origin dating back to the 3rd century BC in the north of China. Blood samples from 47 elder individuals were collected in 2016 and 2017 in the villages of Bekovo, Chelukhoevo, and Novobachaty of Belovo district, and a tiny settlement of Shanda in Guryev District, Kemerovo province.
Figure 1. Approximate location of northeastern part of Altai. Map of Altai-Sayan, showing mtDNA sampling locations.
Sequence analysis
The complete sequencing procedure of modern samples entailed PCR amplification of 2 overlapping mtDNA templates, which were sequenced with Illumina HiSeq 2000 (Illumina Human mtDNA Genome Guide 15037958B). Short reads were mapped using bwa version 0.7.17 (Li and Durbin 2009). All mtDNA genome consensus sequences were called using Unipro UGENE version v40.0 software (Okonechnikov et al. 2012). The haplogroup affiliations reported in this analysis correspond to the current nomenclature of mtDNA based on PhyloTree17 (van Oven and Kayser 2009) and traced for accuracy (van Oven 2015). Variants were scored relative to the RSRS (Behar et al. 2012), with common indels and mutation hotspots at nucleotide positions. Mitochondrial haplogroups were assigned with mitohg v.0.2.8 software (https://github.com/stasundr/gomitohg). As a result, 142 mitochondrial genomes are listed in Table S1, along with corresponding ethnicities, sample locations, and GenBank accession codes. The study provided valuable unique new data from populations and groups that had not been published before.
Data availability
The mitochondrial genome sequence data generated in this study have been deposited in the National Center for Biotechnology Information (GenBank; http://www.ncbi.nlm.nih.gov/Genbank/) under accession numbers MG660498-MG660608.